Superresolution US Breast Cancer Imaging
Development of Motion-Model Ultrasound Localization Microscopy to Support Breast Cancer Diagnosis and Therapy Monitoring in Patients
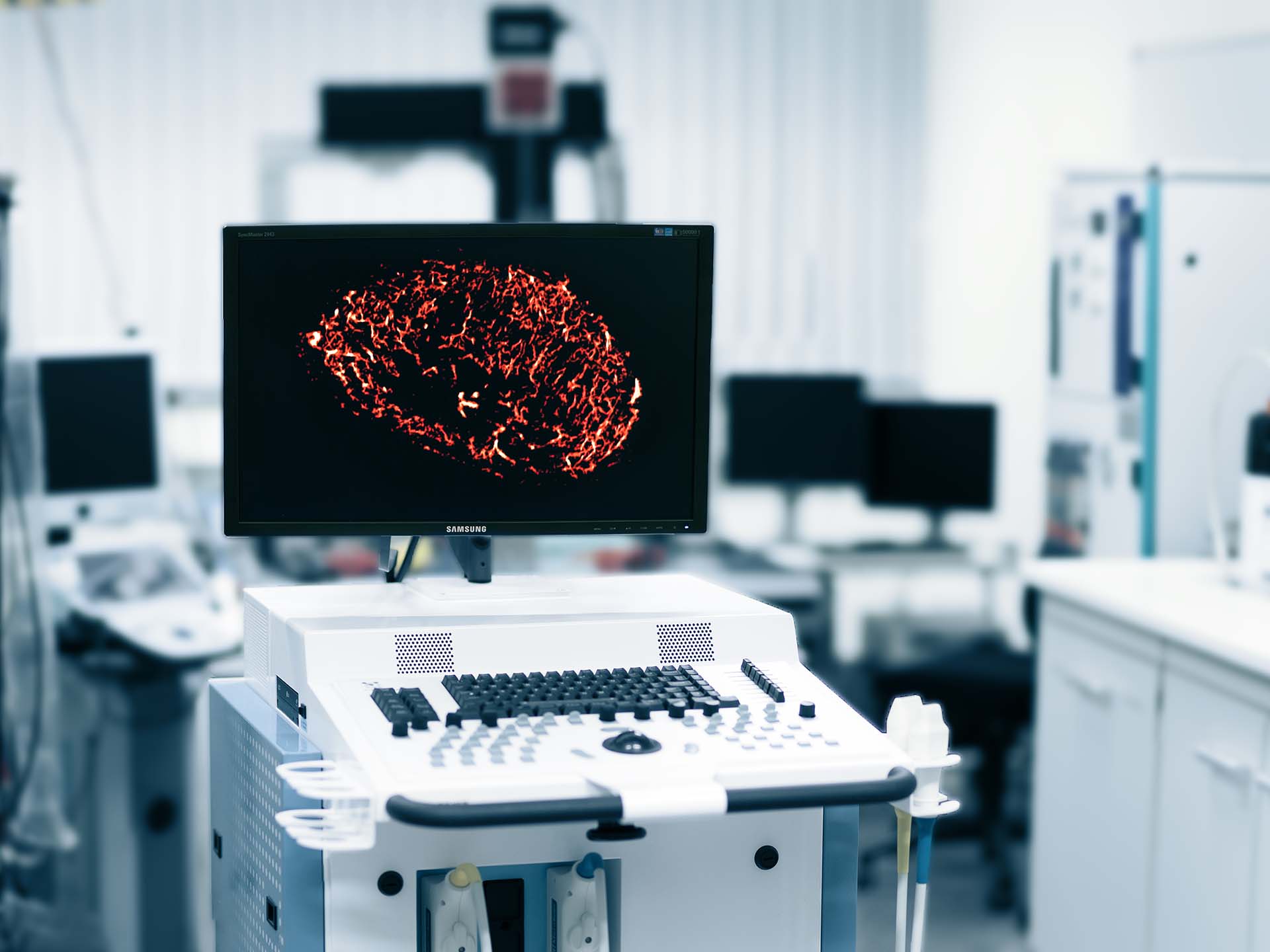
Ultrasound imaging is routinely applied in breast cancer screening and for the dignity assessment of suspect lesions. Supported by the DFG, we developed a super-resolution contrast-enhanced ultrasound method called motion model ultrasound localization microscopy (mULM), which tracks individual microbubbles in video sequences and generates images of the vasculature at a resolution beyond the diffraction limit.
The mULM method enables an accurate assessment of the vascular architecture in tumors, individual blood vessel velocities and flow directions, and the quantification of relative blood volume and perfusion. Previously, we showed in mouse xenograft tumors that this method allows to extract various morphometric parameters and combined with a radiomics analysis supports the automated discrimination of different tumor types.
In this project, we aim to adapt and develop mULM and ultrasound-based radiomics for the application in patients and to advance the technique to 3D acquisition and tracking. In a preliminary analysis of clinical datasets, we were already able to extract super-resolution data of vascular tracks. However, several technical challenges for the robust application in a clinical setting emerged: in particular, patient motion and the lower image resolution together with increased slice thickness and non-optimized injection protocols allowed only to use short image sequences and to extract a reduced number of tracks.
Thus, in an initial step, the mULM methodology will be refined and adapted for its application in breast imaging on clinical ultrasound devices. Subsequently, the 2D mULM method will be applied to monitor breast cancers’ responses to neoadjuvant chemotherapy. Its accuracy will be compared to conventional contrast-enhanced ultrasound data analyses. Furthermore, we strive to advance the method from 2D to full 3D acquisition with a real-time volumetric scanner and compare both approaches in a small cohort of patients with breast tumors of different dignity. To exploit the full potential of mULM data analysis, algorithms will be developed and implemented that allow the automated extraction and clustered analysis of the vascular features. Measurements of similarity as well as cluster analysis will be performed to automatically group the patients. Thus, this project will be a first contribution to the clinical translation of multiparametric super-resolution ultrasound and open the way for ultrasound-based radiomics analysis.